Introduction to Medical Ontology
One of the things that make healthcare a unique domain for big data analytics is the existence of structured medical knowledge, which are often represented as ontologies or knowledge graphs. For historical reason, healthcare and medicine domain have already developed many ontologies for organizing diseases, medical procedures, medications, lab tests, and more. These oncologies give us great resources to understand house care data and to enhance and validate all of the models developed using big data analytics tools. In this lesson we discuss several just ontologies and illustrate how they can help us in our analysis.
Health Data Standards
Now let’s talk about health data standards. There are many different health data standards. In this lecture, we’ll present several important ones. Each has its own unique focus. Let’s illustrate all those data standards through an example of patient encounter. For example, this patient is coming to hospital to get a lab test, and the result of the lab test is stored using LOINC, or Logical Observation Identifiers Names and Codes. LOINC typically represents all different kinds of lab tests.
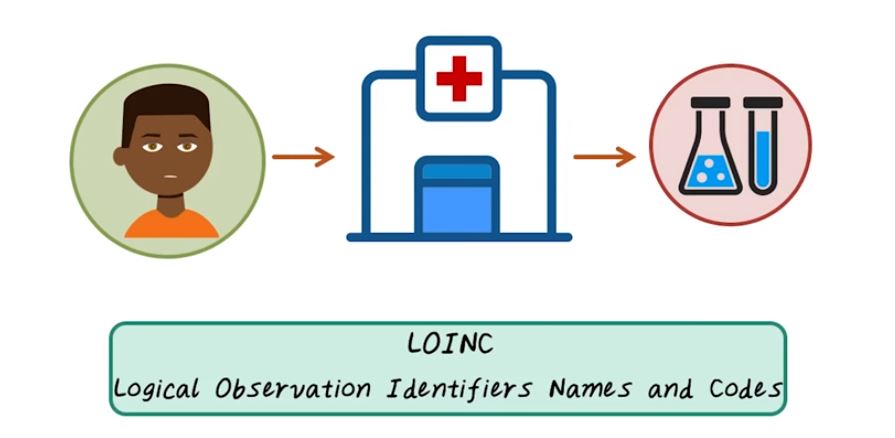
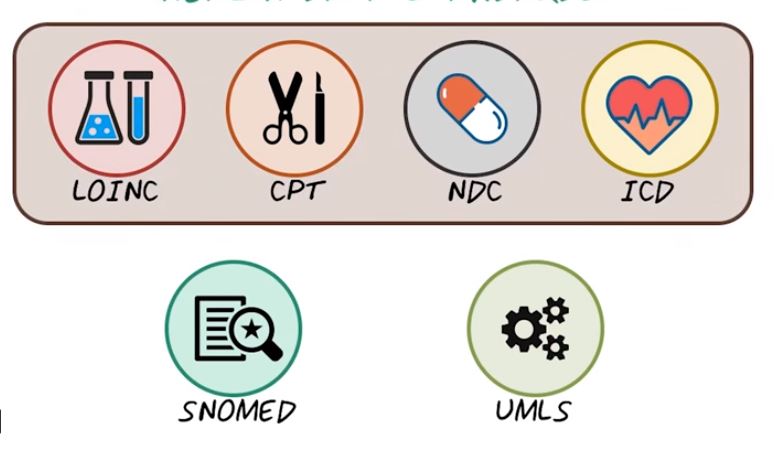
So the lab test result goes to the doctor, and the doctor diagnoses patient with different disease code, or ICD. It stands for International Classification of Disease. That represents different diagnoses and different diseases. Once we have the diagnosis on this given patient, we may want to treat this patient with a medical procedure that’s represented by CPT code, or Current Procedural Terminology. So CPT represents all different procedures that can happen, to give an individual. Of course, the patient can also take some medication, and that’s represented by NDC code, or National Drug Code. So for a given patient,during a typical medical encounter, all different types of information are coded with different health data standards. LOINC code for labs, ICD code for diagnosis, CPT code for procedures, and NDC code for medications, and all those codes interacting with each other can be represented by a medical ontology.

The most popular medical ontology is called SNOMED, it stands for Systemized Nomenclature of Medicine. It’s a huge graph of medical concepts and their relations. Of course, to utilize all this information in a huge oncology like SNOMED, you need software systems to interact with all those concepts and relations. The most popular software for accessing this medical knowledge is UMLS. It stands for Unified Medical Language System. UMLS is a set of software tools that provide integration of multiple sources of medical knowledge and medical ontologies. So next we’ll talk about all these different health data standards. We’ll first cover all those individual set of data standards for different types of medical data. Then we’ll talk about SNOMED and UMLS as a way to integrate all those different medical concepts together. All these health data standards are very important to support common healthcare operations, such as efficient insurance claim processing. For example, when a healthcare encounter has happened and all this information has been recorded through electronic health records, and that information will be sent to an insurance company in order to process those claims so that the doctor can get paid. And all those health data standards support efficient insurance processing. The secondary use of these health data standards are to support research and development. Because house care’s data are encoded largely in structured forms, so that it can be easily analyzed and standardized. Next we’ll introduce all of them in more details.
ICD
First let’s talk about ICD code for diagnosis. ICD stands for International Classification of Diseases which was developed by World Health Organization, and the focus of ICD codes is to categorize diseases.
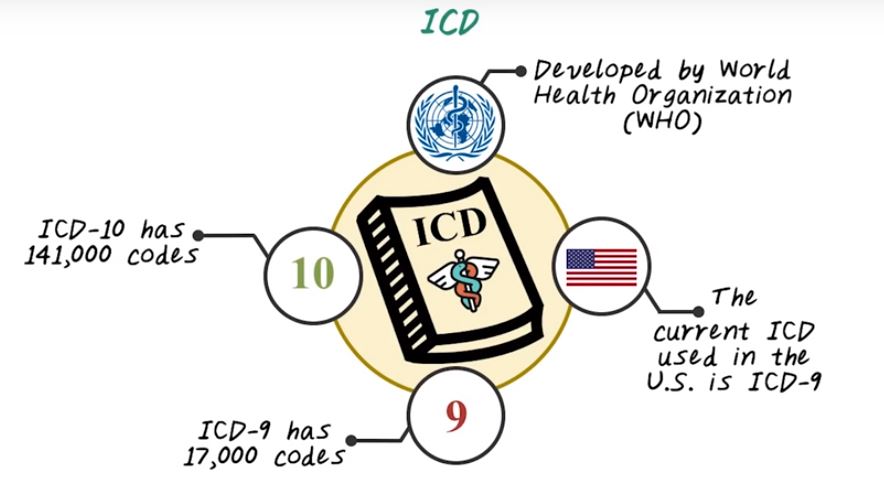
In US currently, we’re using the version nine of ICD and we’re already moving towards ICD-10, and most of the rest of the world are currently using version 10. And version-9 of ICD code has over 17,000 individual codes. It covers both diagnosis and procedures. And ICD-10 code is the next generation of ICD code. It has over 141,000 unique code. Next let’s talk about ICD-9 and ICD-10 in more details. First, let’s introduce ICD-9 code. ICD-9 code has three to five digit with three different levels. Namely, the categories which cover the first three digit, the subcategories which is the fourth digit. And the subclassification, which correspond to the last digit. There are 17 categories plus several supplementary categories in ICD-9. The 17 categories correspond to major disease categories, such as infectious disease, neoplasia, and et cetera.
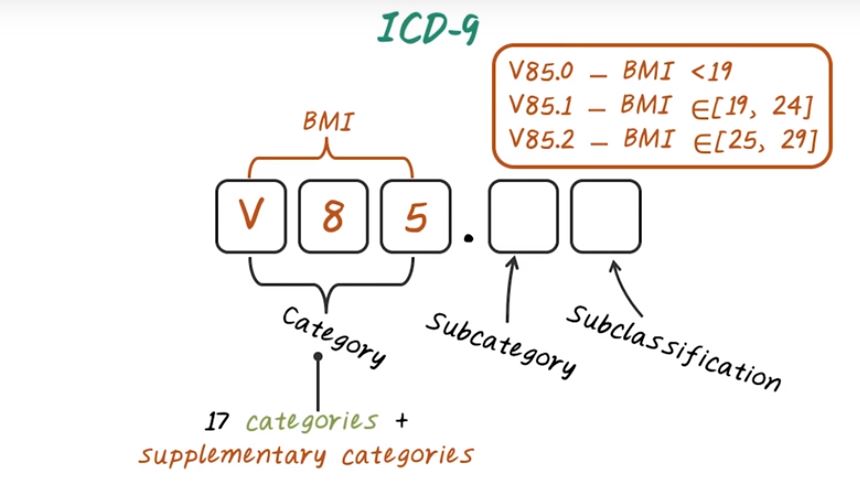
ICD-9 code in the 17 categories have only numerical digits. For example, 250 is a category code for diabetes. And 250.01 is the subclassification for type one diabetes without complications. In addition to the 17 categories, there are supplementary categories starting with letter E or letter V. For example, V85, corresponding to body mass index. And V85.0 indicate BMI less that 19. V85.1 indicate BMI between 19 and 24.
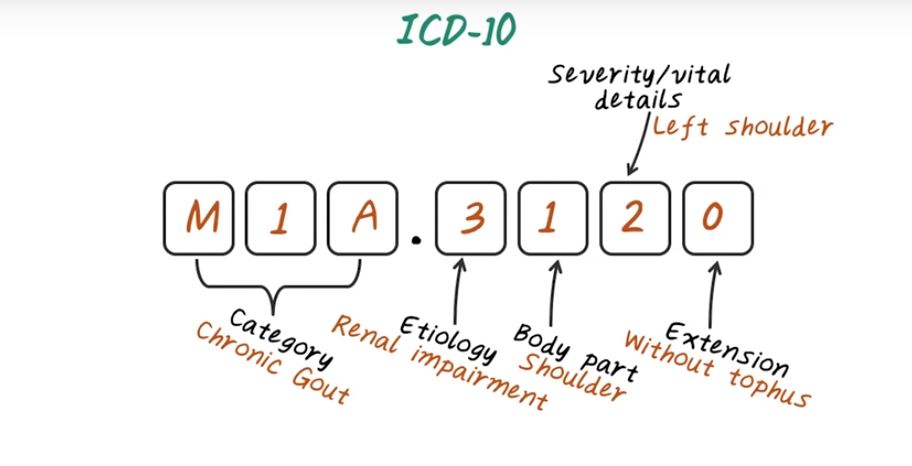
And V85.2 indicate BMI between 25 and Now lets talk about ICD-10 code. ICD-10 code can have seven alphanumerical characters. So it’s longer than ICD-9 code, which only have five digits. First three characters indicate the disease category. The fourth character indicates Etiology of the disease. That is the cause of the disease. The fifth character indicate body part affected. And the six characters indicate the severity of the eonis. And the character seven is the place holder for extension of the code to increase specificity. For example, E110, indicate disease category with type one diabetes. And nine indicate there’s no complication. Here’s an example of a more complicated ICD-10 code. M1A indicates the disease, chronic gout disease. And a three indicate the Etiology of renal impairment. And one indicate the body part, in this case is the shoulder. And two indicate the severity or vital details, so in this case two means the left shoulder. And the last one, zero for extension. Here zero means without tophus. So as you can see ICD-10 code can be quite complicated.
ICD9 – ICD10 Mapping
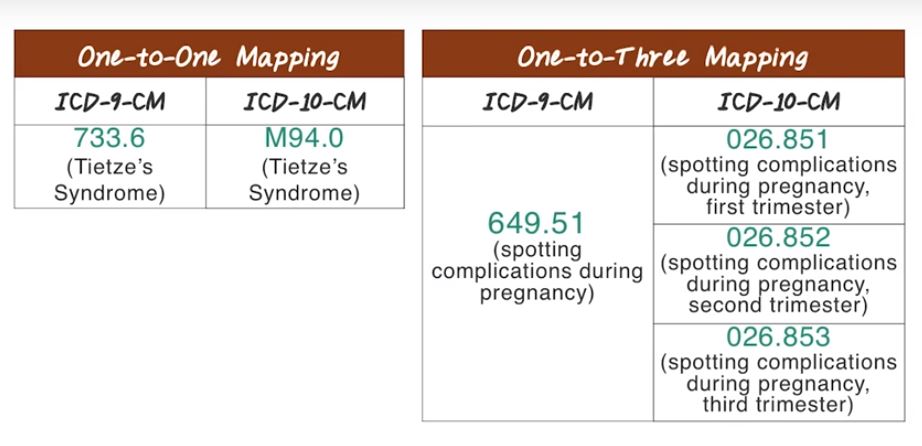
Because of the practical need, there are a huge effort involved in mapping from ICD-9 code to ICD-10 code. In general, a mapping from ICD-9 code to ICD-10 code is a one-to- many relationship, because ICD-10 code is just more specific than ICD-9. So in some cases, ICD-9 is already pretty specific. In this case, they will have one-to-one mapping. For example, for this Tietze’s Syndrome, they are both having unique code in ICD-9 and ICD-10. So the mapping is one-to-one. But in most cases, we’ll see one-to-many mappings. For example, 649.51, spotting complications during pregnancy, maps to three codes in ICD-10. So in this case, complications during each trimester has its separate code in ICD-10. Sometimes the mapping from ICD-9 to ICD-10 can be quite complicated. For example, 733.82 in ICD-9 has a mapping of 2,530 in ICD-10. So ICD 10 is just going through a lot more details about the disease. So, the mapping can be quite tricky.
CPT
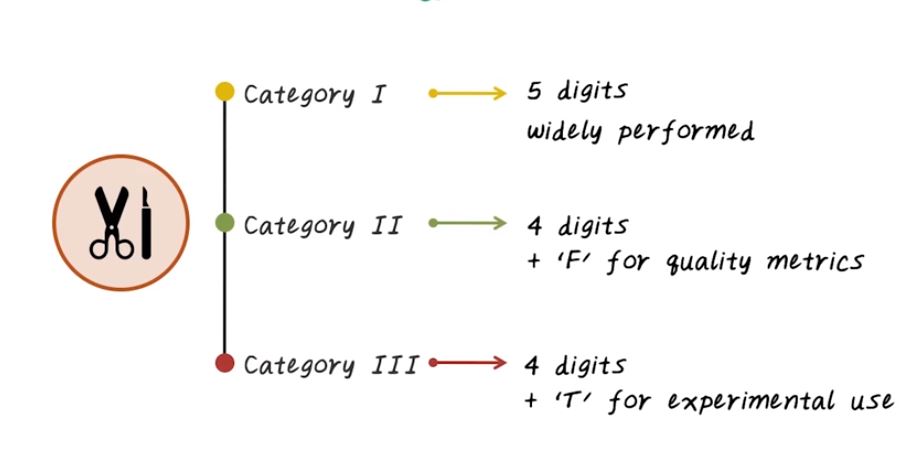
CPT code is another important coding system used in US healthcare. CPT stands for Current Procedure Terminology. The focus of CPT is to describe medical, surgical, and diagnostic services. It’s a US standard for coding medical procedures. CPT is maintained by American Medical Association. And CPT is mainly used by insurance company to determine how much to pay for a medical service. In general, reimbursement rate will be associated with each CPT code has been used in the claims. So CPT is very important in the US, since it tied to how much money doctor will make. Now let’s talk about CPT code in more details. CPT is a five digit code, has three different categories. Category one, corresponding to widely performed procedures. And Category two, corresponding to quality metrics performed in healthcare organization. Then we have Category three, it’s again four digits, follows with letter T for experimental use. Now let’s talk about Category one, CPT code. We can take quick look at the different sections of Category one CPT code. They’re arranged in their numerical ranges. They are divided into six sections, Evaluation and Management, Anesthesia, Surgery, Radiology, Pathology and Laboratory, Medicine. And, here are the Category two CPT code. There are supplementary code for tracking the performance measure. And the different ranges of the code map to different types of services. It include, Composite measures, Patient management, Patient history, Physical examination, Diagnostic/screening process and result, Therapeutic and preventive or other interventions, Follow up or other outcome, Patient safety and Structural measures. For example, blood pressure measured is one of the Composite measures, and there are other Composite measures, as well, they’re all in this range.
LOINC
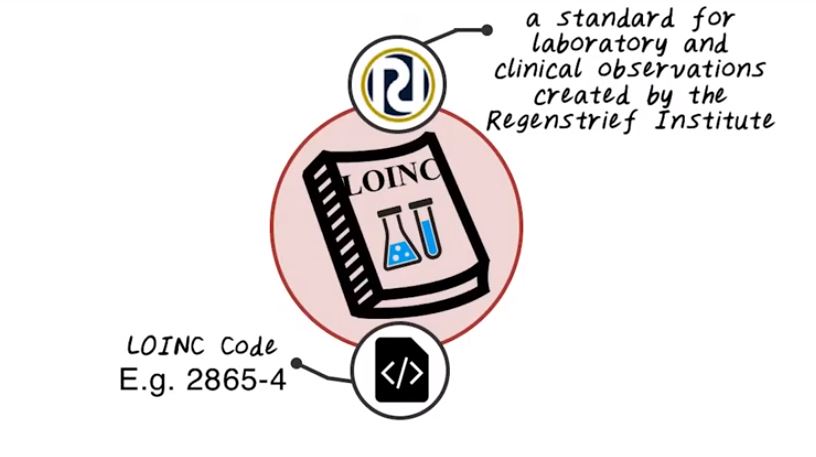
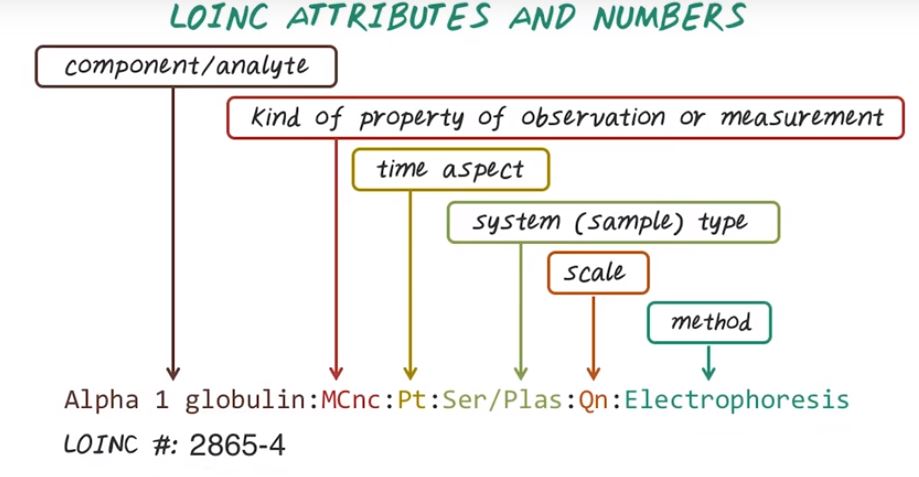
Next let’s talk about LOINC. LOINC is a standard for lab and clinical observation and it’s created by Regenstrief Institute which is a non-profit organization in Indiana. LOINC has been created mainly to capturing lab test. And a LOINC code contains digits like 2865-4, and this is a LOINC code. Of course LOINC code itself has a number and also has attributes associated with this lab test. And different attributes are separated by a column.
Let us talk about loinc attribute and loinc number. Here’s the attribute of a specific loinc code. They’re separated by colon. The first part is the component name and this specify the specific lab test. The second part is the property of the lab measurement. And the third part is a time aspect whether it’s measured at a point of time or over some durations specified here. Here Pt means point of time. The fourth part is a type of sample. For example, serum or plasma in this example. Next is the scale. The scale may be quantitative, ordinal, or nominal or narrative. Finally, we have the method, that has been used to conduct this lab test. And this particular attribute corresponding to a LOINC number of 2865-4.
NDC
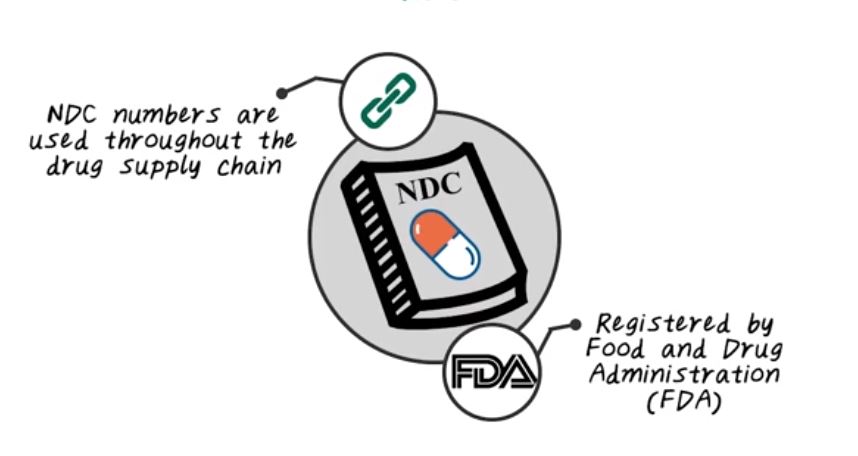
Next, let’s talk about NDC code. NDC stands for National Drug Code. NDC is the standard for medications, and NDC is registered and maintained by FDA, the Food and Drug Administration. And FDA maintains a searchable database of all the NDC code on their website. NDC code are used throughout the entire drug supply chain, from pharmaceutical company to drug distribution companies, to medical community, and to insurance company, and government. They all use NDC code to track medications. NDC have three parts.
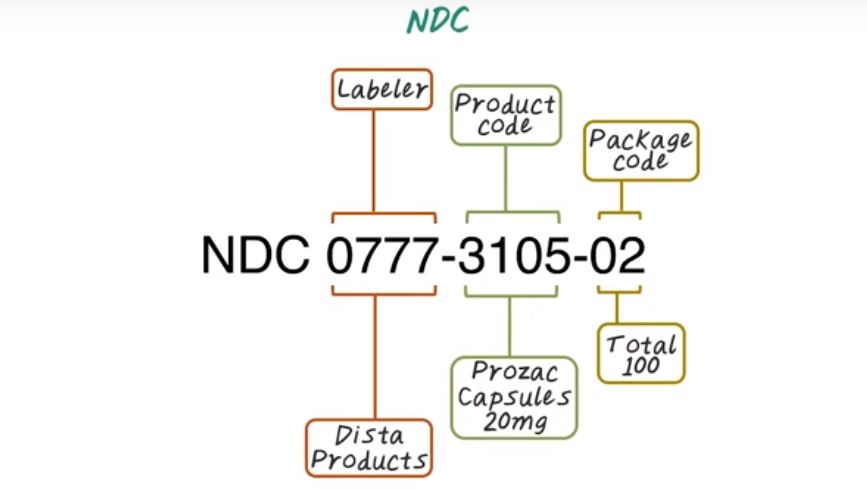
The first part is the four to five digits that indicates the labeler that is a company produce the drug. 0777 is a labeler code for Dista Products, and the second part is the product code to indicate what drug it is. For example, 3105 corresponds to Prozac Capsules of 20mg. And then the final one or two digit corresponding to the package code. In this case, 02 indicate there are hundred pills in this package. NDC code exists in different ways of grouping those three segments. It has a four digit here as labeler, four digit here as the product code, and two digit as a package code. Or it could have a five digit labeler, three digit product code and two digit package. Or it could have five digit labeler, four digit product code and one digit package code. But overall, all NDC code have ten digits.
SNOMED
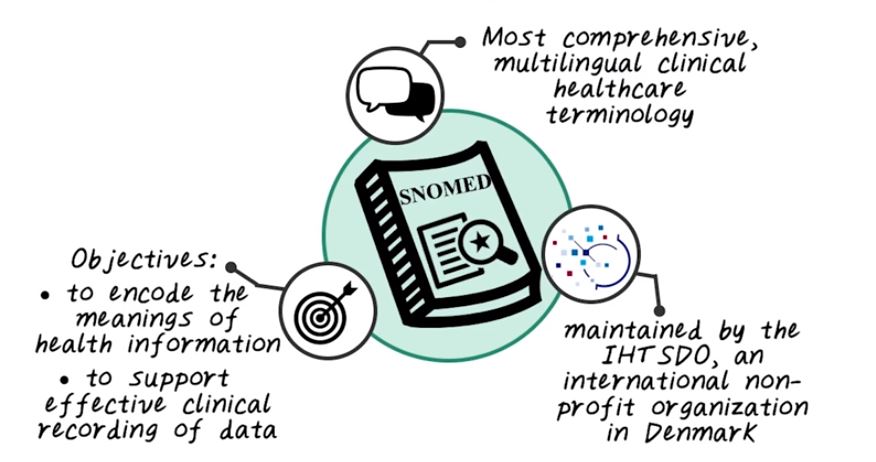
Next, let’s talk about SNOMED. SNOMED is one of the most comprehensive multilingual medical ontology that describes different clinical and healthcare terminologies, and their relationships. And SNOMED stands for systematized nomenclature of medicine. And SNOMED is maintained by another non-profit standard organization called IHT SDO, which is based in Denmark. And the objective of SNOMED is to encode all kinds of health information, and to support effective clinical recording of data was the aim of improving patient care. Next, let’s look into more details of SNOMED.
First, let’s talk about SNOMED Development Cycle. It starts from the central organization, IHTSDO. It maintains the international version of SNOMED ontology. More specifically, it maintains the SNOMED’s development, release, distribution, maintenance and education. And this organization released the SNOMED CT international which is the international version of SNOMED. Then there are different members, those are different countries. Each country can have their own national release. For example, US can have their own SNOMED version. Those different versions are called reference set. There could be SNOMED CT US National Edition and released in 2005. That could be one reference set.
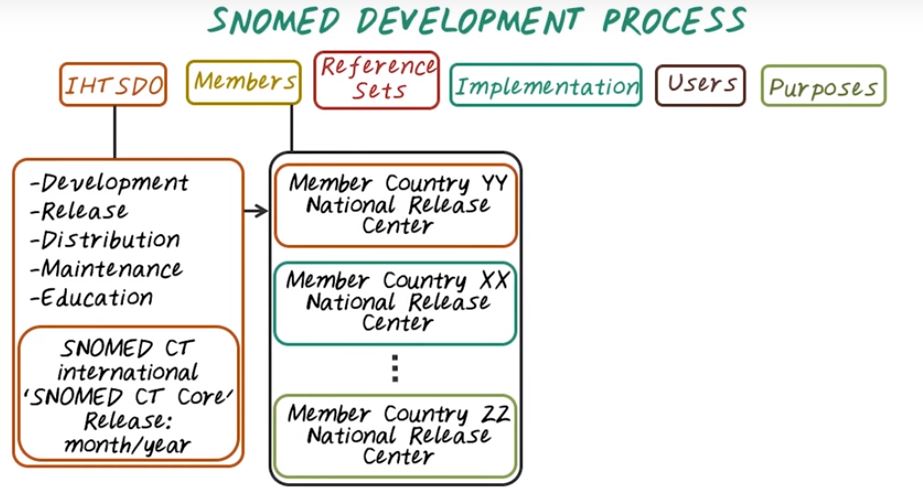
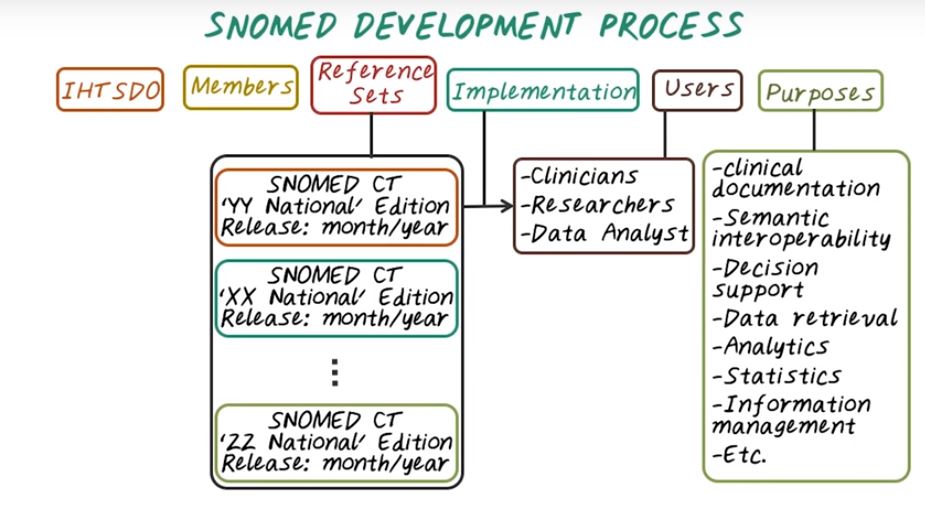
Then there’s different implementation which may only cover a subset of the entire reference set within a country. Then once we have the implementation of SNOMED. And the users of SNOMED are quite broad. They could be clinicians, researchers, or data analysts. And the purpose of SNOMED is to help improve clinical documentation and understand semantic interoperability of medical concepts, and to enable clinical decision support, as well as data retrieval, analytics, statistics, information management purpose.
Logical Model of SNOMED
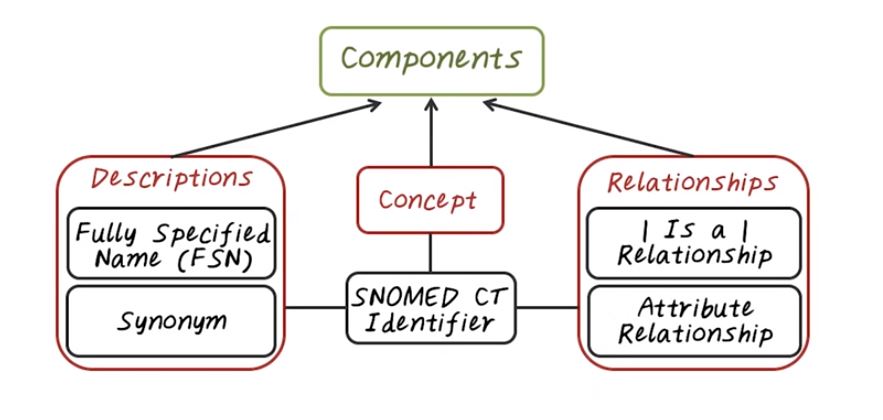
Now lets talk about the Logical Model of Snomed CT. The logical model of Snomed CT is quite simple. It asks three types of components, concepts, descriptions about concepts, and the relationship between concepts. Every concept has an unique identifier, our SNOMED CT identifier. This is a machine readable identifier. Then each concept can be associated with one or more descriptions. And the descriptions provide human readable forms of the concept. There are two types of descriptions. One is the fully specified names, FSN. That is the most precise explanation of that concept. And there are also other synonym that provide different version, or different ways of describing the same concept. And relationship captures interactions between multiple concepts. Usually two concepts. For example, the most important relationship is the is a relationship, or subtype relationship. It gives you a way to generalize a concept from more specific level to more general level. Then there’s the attribute relationship. Each concept can have multiple different attribute.
SNOMED Example
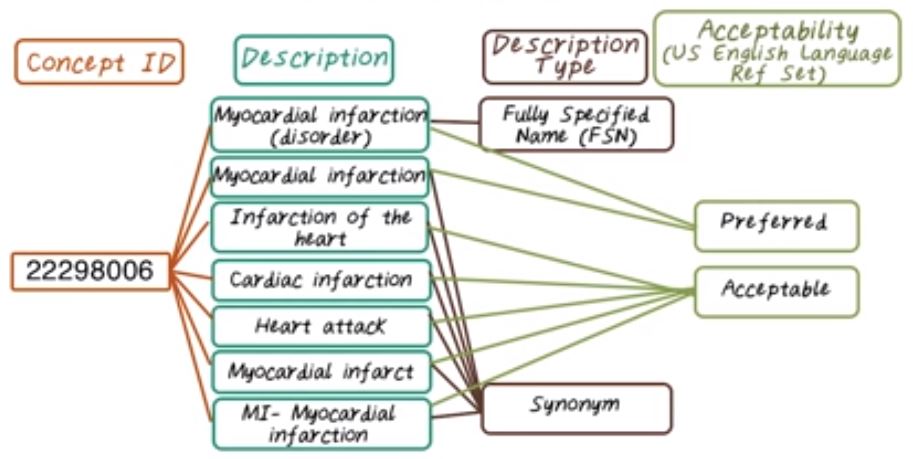
Now let’s see an example of SNOMED concept. The concept ID is the following, 22298006. It is definitely in a machine readable form. This concept ID is associated with different descriptions, and among which myocardial infarction disorder is a fully specified name for this concept. Then there are many synonyms include myocardial infarction, infarction of the heart, MI, heart attack, and so on. Some of those description are considered preferred. For example, myocardial infarction disorder and myocardial infarction. And some are considered acceptable. So the rest of those descriptions are considered acceptable. So note that the concept of preferred or acceptable, they’re implemented in the US English Language Reference Set, which may not be in the reference set from other countries.
SNOMED Relationships
Now let’s talk about IS a relationship. IS a relationship indicate generalization from a specific concept to a more general concept. For example, cellulitis of foot is a cellulitis, so this is a IS a relationship. At the same times, cellulitis of foot is also a disorder of foot. So there’s two different paths to generalize this concept. And you notice that IS a relationship is directional. So two concept are directly linked by IS a relationship. The source concept is said to be the subtype. And the destination concept is said to be the supertype. If we generalize all those concepts to the most general forms, we have a single root of the entire hierarchy. Besides IS a relationship, there are other relationship as well. For example, abscesses of heart Is associated morphology to abscesses. And it has a finding site to heart structure.
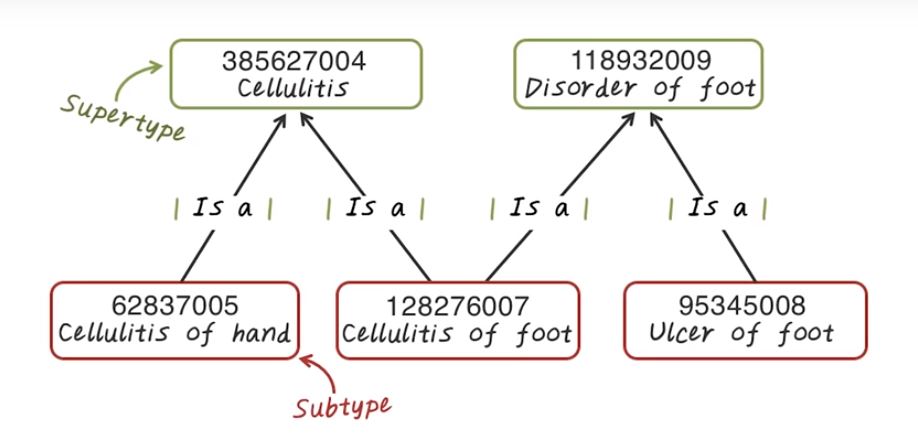
SNOMED Design
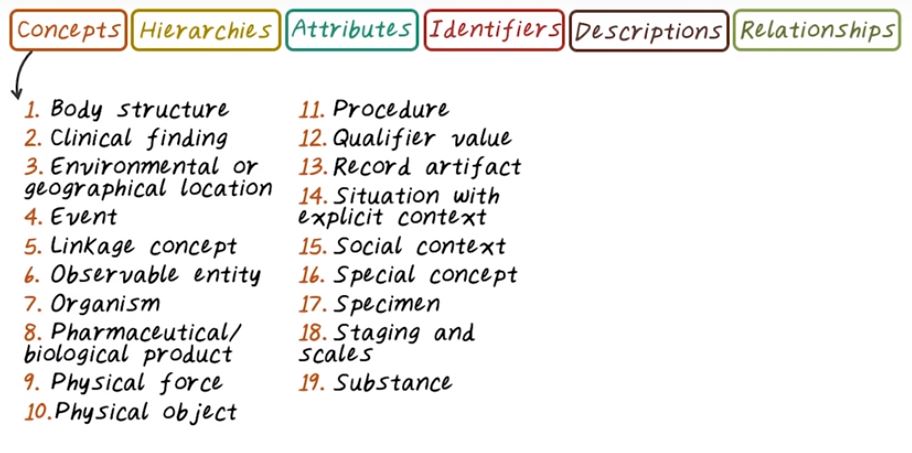
Here’s a summary of SNOMED. SNOMED Ontology consist of different type of concepts, and those concepts are organized in a hierarchy. For example, from top down, there is 19 level of hierarchy, start from body structure go down to the substance. For example, arthritis of knee is the is arthropathy of knee joint is a arthropathy, is a joint finding, and so on, so you can see all those different levels of concepts can be mapped through the hierarchy of the entire SNOMED concept. In the top level are the most general or low granularity concept, and the lowest level here are the high granular or the most specific concepts. Concepts and hierarchies, we also have relationship and their attribute. For example here are two different relationships. Arthropathy is a joint finding, and another one Appendicitis is associated morphology to inflammation. So here associated morphology is an attribute in this relation, and every concept has a unique machine readable identifier and each concept also has associated descriptions. One of those is fully specified name.
UMLS
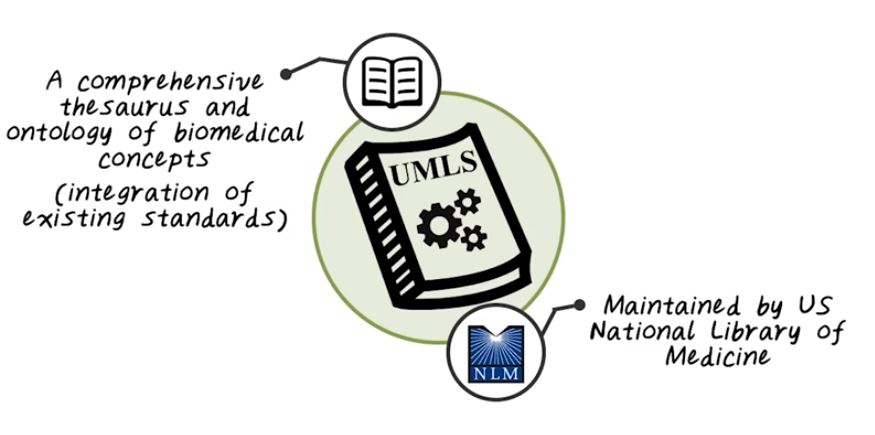
Next let’s talk about UMLS. UMLS stands for Unified Medical Language System. It is a set of software tools that maintained by National Library of Medicine in the US. It’s a comprehensive thesaurus and ontology of all biomedical concepts. It integrates all those existing data standards we just talked about. It also provides software tools to map data to those clinical concepts.
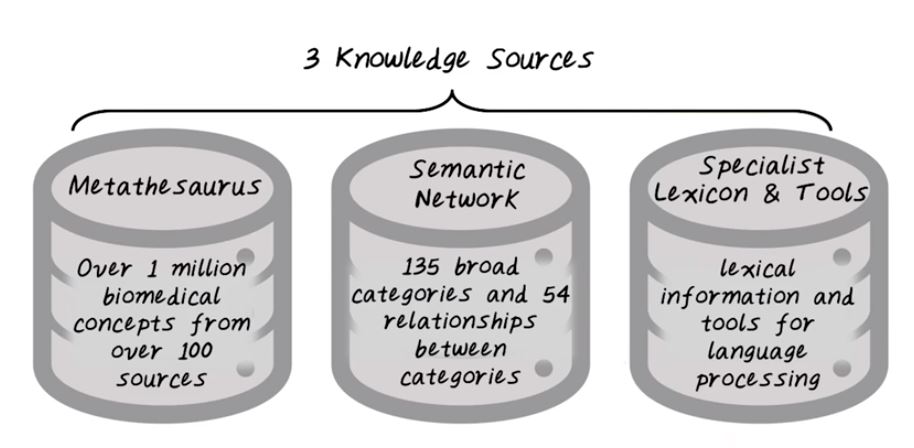
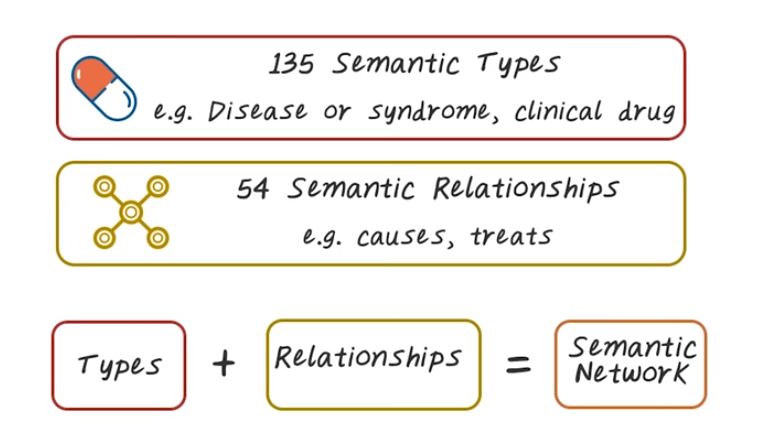
So what are the different components in UMLS? So UMLS recognizes that there are many existing ontologies and terminologies. They want to integrate all of them together, you see one system, so that people can access all of those medical concepts through the systems. There are three knowledge sources in UMLS, the metathesaurus, semantic network, and specialist, lexicon and tools. There’s over 1 million biomedical concepts from over 100 different sources that constitute this medical storage. It includes all the ones we have talked about such as ICD and Snomed. It covers 135 broad categories and 54 different type of relationship between all those concepts, and the semantic type and relationship provide a consistent categorization of concept and their relationship represented in UMLS metathesaurus. Third is the lexicon information and tools that can help processing medical texts. Next, let’s provide more details on each one of them.
Metathesaurus
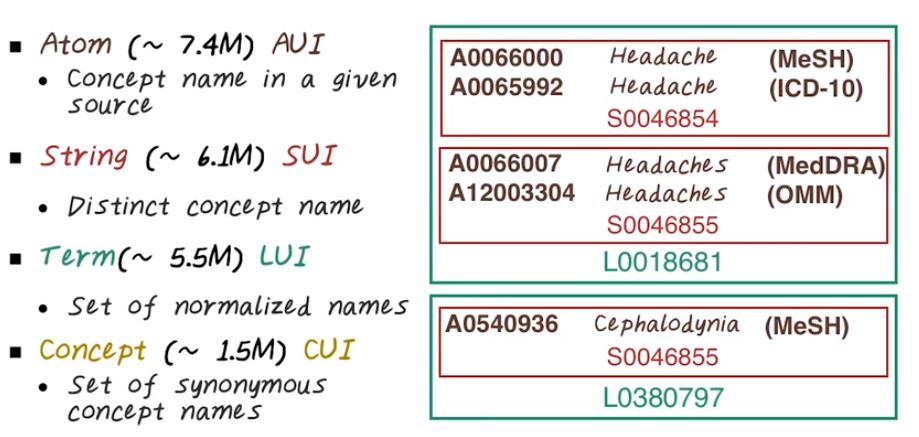
Metathesaurus Concepts. The idea is very similar to SNOMED. Each concept has a specific identifier and organized into a hierarchy. At the lowest level we have atom, at over 7.4 million atoms or AUI. Those are concept name in a specific source. For example, all this different AUIs mapped to something related to headache. Then there are strings, they’re are distinct concept names. So you notice that on the atom level even the same mention can have different AUI because they come from different ontology, different sources. One from MeSH, one from ICD-10. But they have the same string, so string or SUI will be the same. Then we have terms, that’s a set of normalized names. For example here, headache and headaches all map to the same term, or LUI. Then on the highest level, we have concept, or CUI. That’s a set of synonyms. And all of this together, for example, correspond to a single CUI, or cooey.
Semantic Network
Now let’s talk about Semantic Network. So there are over a 135 semantic types such as, disease, syndromes, clinical drugs, all those are different semantic types. Semantic Network organize all those different types into [INAUDIBLE] And then organize those into hierarchies. And there are 54 different type of semantic relationships, such as cause, treat, all those are different type of relationship. And combine them together, the semantic types plus the semantic relationship. That give us the semantic network, and the concept of semantic network in UMLS is very similar to SNOWMED. The only difference here is here we have much richer information because multiple data sources, a different thesaurus, has to be integrated into UMLS, into the same semantic network.
Special Lexicon
Finally, we have the specialist lexicon, which is English language lexicon of common words and biomedical terms. So we have over 300,000 biomedical terms, and their syntax, how the words are put together, morphology, such as inflection, derivation, compounding, all those language expressions, and orthography, that is really the spelling of those terms. And lexicon is used with lexicon tools in a variety of ways for natural language processing For example, MMTX and MetaMap are two software tools provided as part of UMIS to parse medical text